I often use Python to plot data on a map and like to use the Matplotlib Basemap Toolkit. In practice, I use a lot of different libraries to access various data formats (raster, vector, serialized…), select and analyse them, generate, save and visualize outputs, and it’s not always obvious to string one’s favourite tools together into efficient processing chains.
For example, I’ve also become fond of Pandas DataFrames, which offer a great interface to statistical analysis (e.g. the statsmodels module), and its geo-enabled version, GeoPandas. Both Basemap and GeoPandas can deal with the popular (alas!) ESRI Shapefile format, which is what many many (vector) GIS datasets are published in. But they aren’t made for working together. This post is about how I currently go about processing Shapefile data with GeoPandas first and then plotting it on a map using Basemap. I’m using an extremely simple example: a polygon Shapefile of the earth’s glaciated areas from the handy, and free, NaturalEarth Data site. The data is already in geographic coordinates (latitudes/longitudes), with a WGS 84 datum. We therefore don’t have to worry about preprocessing the input with suitable coordinate transforms. (Often, you do have to think about this sort of thing before you get going…). All my code is available in an IPython (or Jupyther) Notebook, which should work with both Python 2 and 3.
So let’s say we have our glacier data in a file called ne_10m_glaciated_areas.shp. GeoPandas can read this file directly:
import geopandas as gp
glaciers = gp.GeoDataFrame.from_file(
'ne_10m_glaciated_areas/ne_10m_glaciated_areas.shp')
glaciers.head()
The output looks something like this:

The geometry column (a GeoSeries) contains Shapely geometries, which is very convenient for further processing. These are either of type Polygon, or MultiPolygon for glaciers with multiple disjoint parts. GeoPandas GeoDataFrames or GeoSeries can be visualized extremely easily. However, for large global datasets, the result may be disappointing:
glaciers.plot()

If we want to focus on a small area of the earth, we have a number of options: we can use Matplotlib to set the x- and y-limits of the plot. Or we can filter the dataset geographically, and only, say, plot those glaciers that intersect a rectangular area in the vicinity of Juneau, AK, that is, the Alaskan Panhandle and the adjacent Western British Columbia. Filtering the dataset first also speeds up plotting, by a lot:
import shapely studyarea = shapely.geometry.box(-136., 56., -130., 60.) ax1 = glaciers[glaciers.geometry.intersects(studyarea)].plot() ax1.set_aspect(2) fig = plt.gcf() fig.set_size_inches(10, 10)

This is remarkable for so few lines of code, but it’s also as far as as we can get with GeoPandas alone. For more sophisticated maps, enter Basemap. The Basemap module offers two major tools:
- a Basemap class that represents a map in a pretty good selection of coordinate systems and is able to transform arbitrary (longitude, latitude) coordinate pairs into the map’s coordinates
- a rich database of country and state borders, water bodies, coast lines, all in multiple spatial resolutions
Features that add on to these include plotting parallels and meridians, scale bars, and reading Shapefiles. But we don’t want to use Basemap to read our Shapefile — we want to plot the selections we’ve already made from the Shapefile on top of it.
The basic syntax is to instantiate a Basemap with whatever options one finds suitable:
mm = Basemap(projection=..., width=..., height=...)
… and then to add whatever other features we want. To transform a (longitude, latitude) coordinate pair, we use mm(lon, lat). The resulting transformed coordinates can then be plotted on the map the usual Matplotlib way, for example via mm.scatter(x, y, size, ...). The code to plot our study area and the city of Juneau, in the Albers Equal Area conical projection (good for high- and low-latitude regions), at intermediate resolution, and including water, ocean, coastlines, country borders etc. is:
from mpl_toolkits.basemap import Basemap
import numpy as np
water = 'lightskyblue'
earth = 'cornsilk'
juneau_lon, juneau_lat = -134.4167, 58.3
fig, ax1 = plt.subplots(figsize=(12, 10))
mm = Basemap(
width=600000, height=400000,
resolution='i',
projection='aea',
ellps='WGS84',
lat_1=55., lat_2=65.,
lat_0=58., lon_0=-134)
coast = mm.drawcoastlines()
rivers = mm.drawrivers(color=water, linewidth=1.5)
continents = mm.fillcontinents(
color=earth,
lake_color=water)
bound= mm.drawmapboundary(fill_color=water)
countries = mm.drawcountries()
merid = mm.drawmeridians(
np.arange(-180, 180, 2),
labels=[False, False, False, True])
parall = mm.drawparallels(
np.arange(0, 80),
labels=[True, True, False, False])
x, y = mm(juneau_lon, juneau_lat)
juneau = mm.scatter(x, y, 80,
label="Juneau", color='red', zorder=10)
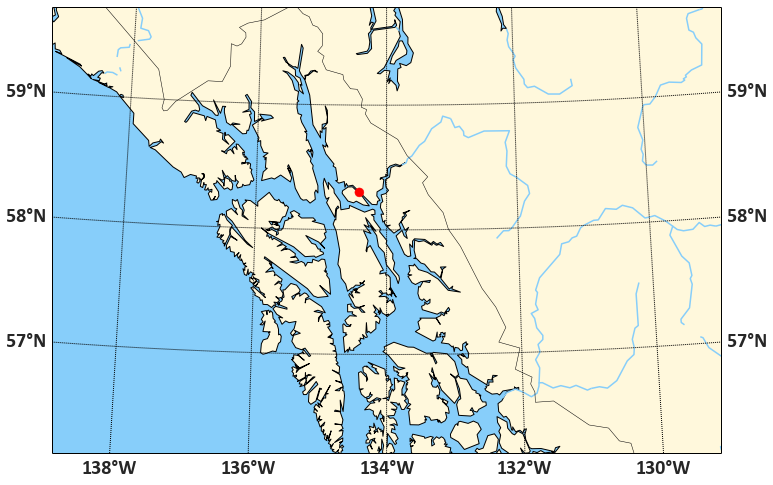
This result may even be quite suitable for publication-quality maps. To add our polygons, we need two more ingredients:
shapely.ops.transformis a function that applies a coordinate transformation (that is, a function that operates on coordinate pairs) to whole Shapely geometries- The Descartes library provides a
PolygonPatchobject suitable to be added to a Matplotlib plot
To put it together, we need to take into account the difference between Polygon and MultiPolygon types:
patches = []
selection = glaciers[glaciers.geometry.intersects(studyarea)]
for poly in selection.geometry:
if poly.geom_type == 'Polygon':
mpoly = shapely.ops.transform(mm, poly)
patches.append(PolygonPatch(mpoly))
elif poly.geom_type == 'MultiPolygon':
for subpoly in poly:
mpoly = shapely.ops.transform(mm, poly)
patches.append(PolygonPatch(mpoly))
else:
print(poly, "is neither a polygon nor a multi-polygon. Skipping it.")
glaciers = ax1.add_collection(
PatchCollection(patches, match_original=True))
The final result, now in high resolution, looks like this:

We could do a lot more — add labels, plot glaciers in different colors, for example. Feel free to play with the code.

